Screening the entire genome for new drug targets for HD
Scientists screen the ENTIRE genome to find new potential therapeutic targets for HD. This ambitious study provides a wealth of data for HD researchers
 By Dr Rachel Harding February 23, 2020 Edited by Dr Jeff Carroll
By Dr Rachel Harding February 23, 2020 Edited by Dr Jeff Carroll
A recently published study in the journal ‘Neuron’ has identified new potential therapeutic targets for the treatment of Huntington’s disease (HD). The work conducted by Professor Myriam Heiman and colleagues used cutting-edge genetic technologies and discovered several genes could modify HD progression in their models in the lab. Many of these genes have not been linked to HD before and could be exciting new targets for researchers to pursue when developing drugs and treatments for the HD patient community.
Ambitious screening of the whole genome
Cells in our bodies contain DNA encoding thousands of genes, each a recipe giving instructions to our cells on how to make a different protein molecule. These recipes are transcribed from our DNA into a message called the messenger RNA. The RNA is then translated by cellular machinery into protein molecules. Scientists can manipulate these processes in the lab to understand the role of different genes in our bodies.
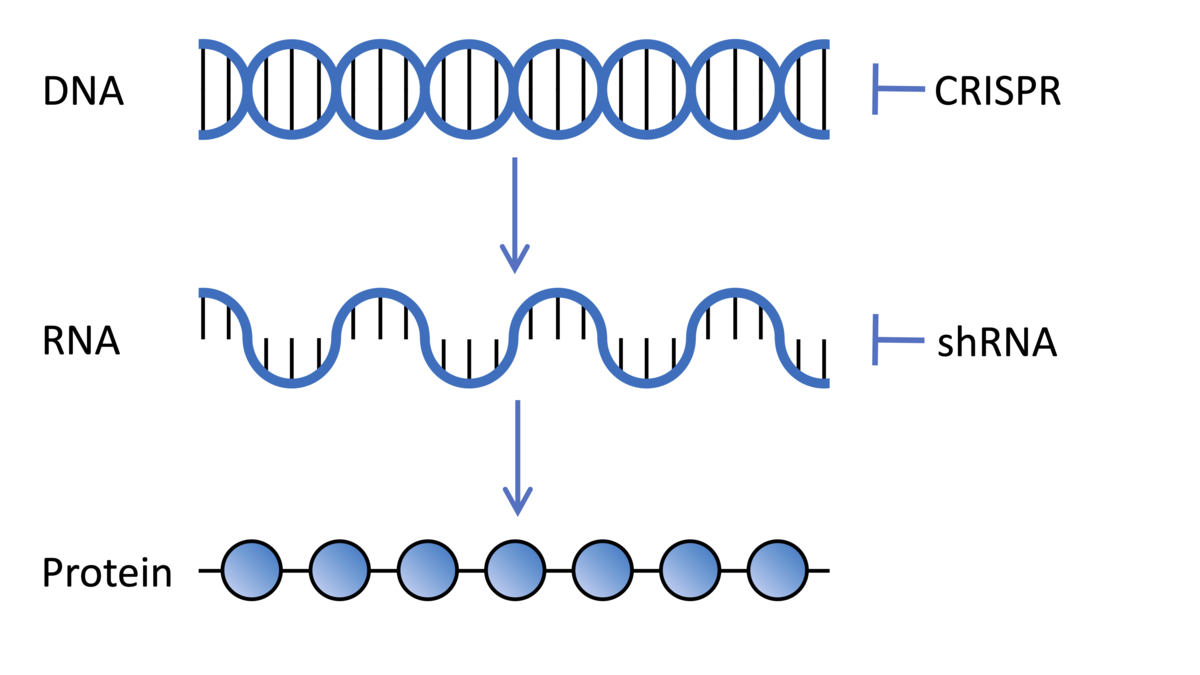
Genetic screens look to understand the role of a single gene in different contexts, in this case, the researchers were interested as to the role of all the different genes in our cells in protecting against the damaging effects of the HD mutation. So the idea is to mess with every single gene in turn, to try and discover whether that gene has any impact on HD symptoms.
Genetic screen technologies can work in lots of different ways, but they all aim to stop or lower the expression of proteins from specific genes. Genes may be targeted directly by editing the genome itself. Other technologies interfere with the message RNA which is transcribed from the gene and is essential for the cells to make the protein which the gene encodes.
This might sound familiar to HDBuzz readers as these are similar technologies to those being used in huntingtin-lowering therapies which are currently being assessed in various clinical trials. Whereas these huntingtin-lowering therapies target just the huntingtin gene, in the case of this genetic screen, the researchers are targeting every gene in the genome, one-by-one, to work out the role they play in HD.
The Broad Institute, where Prof. Heiman is based, is a world leader in developing libraries which can be used for genetic screens. In this study the researchers used two different technologies in their screen, both of which are delivered into brain cells by special types of viruses. Firstly, short-hairpin RNAs which target the messenger RNA and turn down expression of the gene by interrupting the message from being translated into the functional protein molecule. Secondly, CRISPR was used to directly edit the gene sequence in the genome, disrupting its ability to be switched on to make the protein it encodes.
Systematic genetic screens in different animal models have been around for decades for more simple systems such as worms and flies. However, these types of experiments have been much more technically challenging to perform for the brains of mammals which has been a hindrance to scientists interested in conducting these screens to understand different neurodegenerative diseases.
Prof. Heiman’s team were able to overcome these difficulties by finding a way to pool and concentrate the reagents which need to be injected into the brains of the mice in the genetic screen, and were able to directly target the striatum which is the area of the brain they were interested in studying. The striatum is the most heavily impacted brain region in HD patients, which is why this area was chosen.
20,000+ genes were investigated in this one study
Rather than look at familiar genes associated with neurodegeneration in their mouse models, the scientists in this study took an unbiased approach and completed a genome-wide screen to look at the role played by almost every gene. In fact, they screened nearly all of the approximately 22,000 genes found in mice! This was an incredibly ambitious approach and provides a wealth of data to researchers in the HD field and beyond.
As this was the first systematic screen of all genes in the mammalian central nervous system, the researchers used normal mice with no known mutations to work out which genes are important in brain cell survival in normal conditions. Genes which had been previously identified in systematic screens in more simple models such as flies and worms were shown to be important in mice too.
In this study however, many new genes were identified including several which play a role in metabolism in cells. These were not previously identified in other systematic screens in flies or worms, which is probably because the mammalian central nervous system requires more energy and is more dependent on genes which help the cells make energy. These findings are a good reminder of how important it is for scientists to consider their research findings in the context of the animal models being investigated.

In addition to the control mouse model, two different HD mouse models were used in this experiment, R6/2 and zQ175, both of which are extensively described in the HD research literature. By comparing the genes identified in the screen in the HD mouse models to those identified in the control mice, scientists could work out which genes were specifically important for HD, rather than genes which affect brain cell function more generally.
For the genetic screens conducted on the two HD mouse models used in the study, approximately 500 genes were identified as being important in HD progression. Many of these genes play roles in pathways scientists have previously identified in other studies such as the genome-wide association studies (GWAS) which looked for genes which can alter the age-of-onset of HD symptoms in human patients. These include genes involved with DNA damage repair pathways which maintain the integrity of our genetic material as well as genes in transcription pathways which regulate how the messenger RNA is processed in cells and therefore which protein molecules are made.
New gene targets were identified in the screen too, including genes belonging to the Nme family. Nme genes have been previously reported to be linked to spread in some cancers but this is the first time they have been connected to HD. Heiman and colleagues think that targeting the Nme pathway may be important in helping the brain cells get rid of mutant huntingtin protein in HD brains. If we can design therapeutics which modulate this pathway, this could be a potential way to help treat HD.
New leads for making new HD medicines
Even with lots of ground-breaking clinical trials underway testing different therapies for HD patients, it is important that researchers continue to look for alternative ways to potentially make new medicines for HD. This research provides a wealth of data on HD as it works in mouse model brains and also gives us ideas of new targets to pursue as potential drug targets, which may one day end up in the drug discovery pipeline. It will be exciting to see how these new leads are followed up by researchers around the world and also how this technology might be applied to other neurodegenerative diseases.


