Meet the enemy: neutron ray reveals HD protein structure
Scientists use beams of neutrons to study the structure of clumps of mutant huntingtin protein
 By Dr Jeff Carroll May 26, 2011 Edited by Professor Ed Wild
By Dr Jeff Carroll May 26, 2011 Edited by Professor Ed Wild
The mutant huntingtin protein forms clumps, or aggregates, in brain cells. Many scientists believe these clumps contribute to the death of these cells and symptoms in HD. Now scientists have used a beam of neutrons to study the earliest structures formed in these aggregates.
Huntington’s, Huntington, Huntingtin…
The words used to describe Huntington’s Disease can be confusing. HD is caused by mutation in a gene confusingly called “huntingtin”. Genes are used, like blueprints, in cells to make proteins. Proteins are used as structural elements in your cells, and also serve as small machines that conduct most of the business of the cell.
So the huntingtin gene is the blueprint for the huntingtin protein. It’s the huntingtin protein that actually causes HD, not the genetic mutation itself. The basic job of researchers working in HD today is to understand how the mutant huntingtin protein causes HD symptoms.
Aggregates in the brain
In 1997 two groups of scientists provided evidence that the huntingtin protein has a tendency to stick to itself, forming clumps of protein. A team lead by Dr Erich Wanker in Berlin found that purified huntingtin protein in a test tube forms clumps (or ‘aggregates’). Interestingly, this only happened when the huntingtin protein was mutated, as it is in the disease.
At the same time, a group led by Drs Marian DiFiglia and Neil Aronin at the Massachusetts General Hospital in Boston described very similar aggregates in the brains of human patients who had died from HD - suggesting that the clumping process may be relevant to the disease.
In the 14 years since these pivotal discoveries, there has been a lot of debate among researchers about what these aggregates do in Huntington’s disease. And not just HD - clumps of protein are also found in patients with other neurodegenerative diseases including Alzheimer’s and Parkinson’s Disease.
Some scientists believe these aggregates are simply the by-product of a dying brain cell. Others believe that these aggregates themselves kill brain cells and cause disease - and so are worth studying in great detail. These scientists believe that understanding the first step of the 'aggregation’ process will teach us about how the mutant huntingtin protein kills cells.
A neutron ray? Seriously?
“These kinds of experiments are excellent for solving scientific debates about the nature of huntingtin aggregation, but there’s a long way between these basic scientific observations and new therapies for HD ”
Studying the process of huntingtin aggregation is very difficult, because individual protein molecules are very small. They’re so small that traditional microscopes don’t have enough power to see them.
Enter neutrons. Neutrons are tiny sub-atomic particles usually found in the middle of atoms - one of the tiniest building blocks of matter. Beams of neutrons can be produced from certain nuclear reactions, and these beams can be passed through a sample. The material in the sample will ‘scatter’ the beam - kind of like a flashlight shining through a glass of water.
By studying patterns in the scattered beam, scientists can calculate what the material looks like. This is a bit like deducing the shape of someone by examining their shadow. It’s useful, because beams of neutrons are able to create very detailed shadows from very small objects - like proteins.
But nuclear-reaction fueled neutron beams aren’t exactly easy to find - so how could we use them to study HD? A team lead by Dr Valerie Berthelier was able to access a neutron beam source at Oak Ridge National Laboratory in Tennessee. It’s the most powerful neutron beam in the United States.
To study HD aggregation in great detail, Berthelier’s team made lots of a small piece of the huntingtin protein - the piece with the mutation that causes Huntington’s disease. This purified huntingtin protein was placed in the pathway of the neutron beam, and the aggregation process was watched in real time.
So, how does aggregation happen?
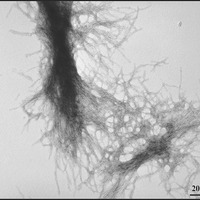
Berthelier’s team showed that purified huntingtin protein clumps together very quickly, as other researchers have shown before. Within a few hours, all the protein was bound together in clumpy aggregates. They then used a different technique - called electron microscopy to examine the aggregates, which looked like the expected fibrous material - kind of like strands of rope.

The advantage of the neutron technique is that it allowed the aggregates to be followed from a very small size. They estimate that two to three individual huntingtin protein molecules clump together to form a small sphere. These spheres quickly grew into long ropes made from fibres of huntingtin protein, twisted together.
Limitations and conclusions
The huntingtin protein is very large - it’s more than six times larger than the average protein in a human cell. That makes it difficult to study, because large proteins are hard to handle in the laboratory.
Because of these difficulties, many scientists work on small pieces of the whole huntingtin protein. The experiments described here used a small fragment that is actually only about the first 1% of the huntingtin protein. Working on a fragment like this deliberately ignores the more complex things that happen in cells, where the whole protein is present, along with many other proteins.
Also, because these measurements are so difficult, a very large amount of huntingtin protein is required - much more than would normally be found in cells. It’s not clear if the things observed in this study are the same as what happens in cells, when much less huntingtin is around.
These kinds of experiments are excellent for helping to solve scientific debates about the nature of huntingtin aggregation, and it’s exciting to see advanced technologies like this applied to the study of HD. Because of the limitations described, though, there is a long way between these basic scientific observations and new therapies for HD.


