Focusing in on fibrils; scientists give us a glimpse of huntingtin protein clumps
Scientists have used powerful microscopes to look at sticky fragments of the Huntington’s disease protein, shedding light on these structures which are thought to drive the disease

A group of scientists from the EPFL in Lausanne, Switzerland have published a paper in the Journal of the American Chemical Society, describing clumps made up of a fragment of the huntingtin protein. A word that’s commonly used to describe these is “aggregates.” Using very powerful microscopes, the team was able to zoom in and look closely at the details of the 3D structures of these samples. The build-up of huntingtin protein aggregates is thought to be an important feature of Huntington’s disease (HD), contributing to the progression of the disease. But until recently we knew very little about what they looked like. With these exciting new glimpses of aggregates under the microscope, scientists hope to build tools to visualize them in the brains of people with HD, or even send harmful aggregates to the trash can in brain cells.
Aggregates, amyloids and fibres – what does this all mean?
Many diseases affecting the brain, including neurodegenerative diseases like Parkinson’s, Alzheimer’s and Huntington’s, are characterised by the build up of clumps of protein molecules in brain cells. In HD, it is a small and sticky fragment of the huntingtin protein itself that forms these clumps, which scientists often refer to as huntingtin aggregates.

“Aggregate” is a fancy word for when lots of copies of the same protein molecule stick together to make much larger three-dimensional structures. Sometimes these aggregates are a jumbled mess of lots of protein molecules all higgledy piggledy. But other times, the molecules are much more organised and form repetitive structures. Some of these more organised structures look like fibres and are called amyloids or fibrils.
You can think of these different organisations of protein molecules like a tower of Jenga bricks. Each brick represents a single protein molecule. When the bricks are all stacked neatly together into a tower, this looks a bit like protein amyloids or fibrils. But when the bricks eventually fall down into a messy pile, this is more similar to what we think a disorganised protein aggregate might look like.
Scientists are generally (and annoyingly) lovers of jargon so you will see that they often use all these words interchangeably. But for the purposes of this article, we are going to be focussed on huntingtin fibrils; organised three-dimensional fibres made up of lots and lots of copies of a small and sticky fragment of the huntingtin protein.
Of mice and men… and bacteria
Aggregation of the huntingtin protein is a long-documented feature of Huntington’s disease. In brains from people who have passed from HD, we can use dyes and other nifty chemical labels to see these aggregates under the microscope in different types of nerve cells. In animal models of HD, which are genetically engineered to make the small sticky fragment of the huntingtin protein, scientists have shown that these aggregates accumulate over time. In many HD model animals, the level of aggregates in different parts of the brain are associated with the severity of HD-like symptoms.
“Bacteria are engineered by the scientists to be huntingtin protein factories, making lots and lots of copies of this molecule”
One of the problems with looking at the aggregates in the brain is that there are lots of other molecules in the cells where we find aggregates, so we generally have to use special stains which stick to the aggregates to see them. However, this approach doesn’t give us very detailed insight into the types of aggregates present or their 3D structures.
To overcome this problem, scientists look at highly pure samples of aggregates which they make synthetically in the lab. Harmless bacteria are engineered by the scientists to be huntingtin protein factories, making lots and lots of copies of this molecule. The scientists can then fish out huntingtin from the bacteria and use these samples to make fibrils in a test tube which look similar to those we see in people. The fibrils can be made with unexpanded huntingtin protein or expanded huntingtin, corresponding to the huntingtin protein with and without the HD mutation. This means that scientists can investigate the effects of the HD mutation on the fibrils.
What can mighty microscopes reveal about these aggregates?
After making these synthetic huntingtin fibril samples, the team of researchers from Switzerland looked at them using a fancy piece of equipment called a cryogenic electron microscope. This type of microscope allows you to really zoom in and see the fibrils in lots of detail. The fibrils are extremely small – only 3-10 nanometers across, about 100,000 times smaller than the thickness of your fingernails – but are easily visible under this type of microscope.
In this study, the scientists took lots of pictures of the fibrils using the microscope and then used special software to average together similar looking images. This averaging process improves the quality of the image, which makes the features of the fibrils easier to see – a bit like changing the contrast or brightness on your phone screen to see the display more clearly.
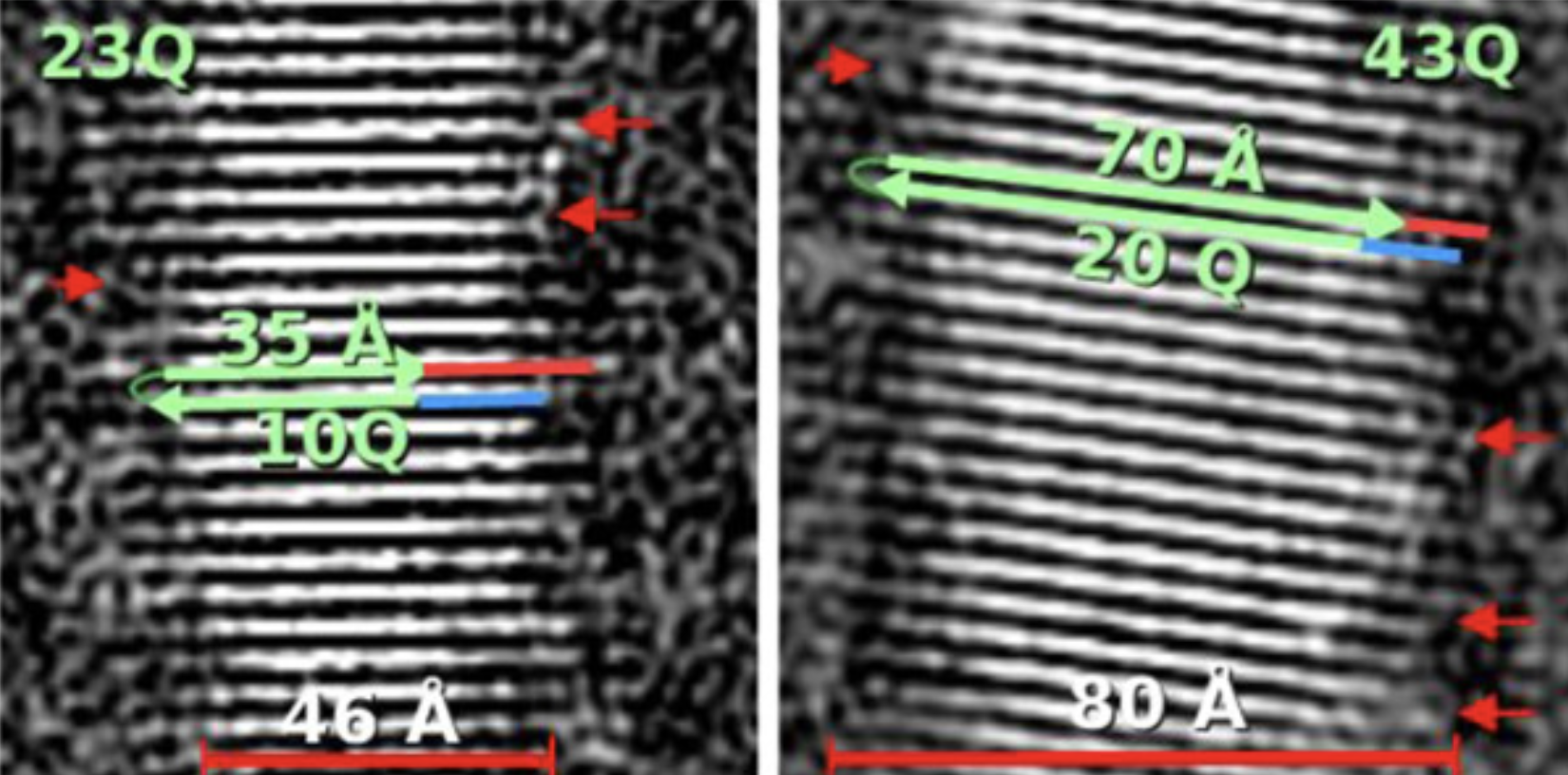
From these images of the fibrils, the scientists were able to measure their dimensions and work out how all the huntingtin protein molecules were organised. They could see that they were stacked together and lined up into flat ribbons, looking a bit like if you took lots of Jenga bricks and lined them all up next to each other to make a thin, single layer of bricks. Many ribbons of huntingtin are layered on top of each other in the fibrils, which would be as though you added more and more layers of lined up Jenga bricks on top of the first.
Interestingly, the researchers found that the HD mutation led to changes in the dimensions of the huntingtin protein fibrils, as well as changes in the number of ribbons of huntingtin stacked on top of each other. The scientists also made fibrils from an even smaller fragment of the huntingtin protein which is missing a region right at the beginning of the molecule. They showed that these fibrils were much more disorganised and were made up of a mixture of different organisations of the huntingtin protein molecules.
These findings are important because they show that the Huntington’s Disease mutation and other regions of the huntingtin gene affect the 3D structure and organization of huntingtin protein fibrils. Fibrils which are uniform of more disorganised, might gum up the works in different ways so this is important to understand.
How will this work help people affected by Huntington’s disease?
Our in-depth understanding of the structure of aggregates in the Huntington’s disease brain is still somewhat in its infancy but we can look to work in other disease areas to see what promise this type of study can hold (beyond generating really cool images of the fibrils of course).
“The Huntington’s Disease mutation affects the 3D structure and organization of huntingtin protein fibrils”
In the field of Alzheimer’s disease research, this type of approach is now being used to look at fibrils from the brains of patients who have passed. This work has revealed an astonishing level of detail of the fibril structures, showing precisely where each atom is located. Comparing fibrils from people with different forms of Alzheimer’s disease, scientists could see subtle differences in their organisation and showed that there are differences among patients, animal models of Alzhiemer’s disease, and the synthetic fibrils generated in the lab. For other types of fibrils scientists have examined, the variation from patient to patient is significant, although it is not yet clear how this relates to symptoms or disease severity.
Other studies show how brain imaging molecules called PET ligands bind to the fibrils. The Huntington’s field has a PET ligand which binds to fibrils (we wrote about this recently on HDBuzz) but we don’t yet know exactly where it binds on these structures, so maybe one day scientists will be able to use this approach to better understand the PET ligand.
Overall, the work by the researchers at the EPFL is an exciting step forward as we begin to understand more about huntingtin fibrils and has laid a foundation for future studies where we might glean more information about this important feature of HD.
Learn more
For more information about our disclosure policy see our FAQ…