
Huntington’s disease therapeutics conference 2023 – Day 1
Check out research updates from Day 1 of the 2022 HD Therapeutics Conference #HDTC2023


Hello from Dubrovnik, Croatia, where the 2023 CHDI Therapeutics Conference will be taking place from Monday, April 24th, through Thursday, April 27th!
This conference is a big one for HD researchers worldwide, from industry, academia, and nonprofit. Dozens of scientists will give talks on all things HD, from genetics, to therapeutics, to clinical trial news.
The HDBuzz Editorial team will be on the scene starting on the morning of Tuesday, April 24th, live-tweeting scientific talks and updates on the progress of clinical trials. Our Twitter updates are compiled below. Continue to follow live updates for the rest of the conference with the hashtag #HDTC2023.
For a summary of last year’s conference, start here: https://en.hdbuzz.net/320 We’ll post summaries in article format for each day of the conference.
Knowing what we need to know
The first talk of the morning comes from Dr. Vahri Beaumont from CHDI, who will give an overview of what we still don’t understand about HD and what we need to know to better develop therapeutics. She is first discussing the history of our understanding of HD genetics and brain changes, from CAG repeats, to loss of brain cells and circuits, which scientists have come to understand through the study of human brain tissue donations and brain imaging.
We have known for a while now that people with HD who have the same CAG number can start to get symptoms at different ages. One reason for this is other genetic differences in a person’s DNA code. Scientists are studying these DNA letter changes to better understand how they might alter HD onset and might also be exploited for making new medicines for HD. Many of these other genetic differences affect “somatic instability,” in which the CAG mutation that causes HD mutates even further in some brain cells, getting even longer. Long CAG repeats in the huntingtin gene leads to an expanded huntingtin protein, which over time can be toxic to different parts of brain cells. Vahri reminds us that there is still a lot we don’t understand about the precise sequence of events which link the HTT gene expansion to the symptoms people with HD experience.
For example, we are still unclear on exactly which HTT gene product is the key player in disease – is it the message molecule? The protein? Protein clumps? Perhaps they all play a role in HD. Another question that remains unanswered is whether the “bad” copy of huntingtin is messing things up, or whether the loss of one “good” copy leaves brain cells without some function. Regardless of these questions, several therapeutic approaches that target HD genetics are already being tested in the clinic. Some focus on total huntingtin, both normal and expanded; others target only the expanded form.
The good news is that there are many different companies testing out all kinds of approaches in the clinic, testing many different hypotheses. Perhaps a combination of these therapies may be the best way to treat HD. Thanks to the generosity of brain donations from people with HD after they pass, scientists are continuing to make breakthroughs to understand the disease in people using these very precious tissue samples.
Scientists are using the most cutting-edge technology to understand what is happening with huntingtin messages and protein in different types of cells and why certain types might be more vulnerable. Using many different kinds of animal models, researchers are building a better picture of what happens inside the brain during HD and how we might intervene. Animal models also allow us to test interventions like drugs at very early stages of HD.
Vahri points out that there are some limitations to mouse models which don’t show all the symptoms of HD in people. Scientists are continuing to develop and use multiple models so they might best test drugs before they are used in people in the clinic.
One of the biggest goals in HD research is to be able to start treatment before symptom onset. This is not easy, but a very strategic staging system, the HD-ISS https://en.hdbuzz.net/325, will help scientists achieve this.
Data Sharing
Dr. David Howland from CHDI is introducing the first official data sharing session of the conference. It will focus on huntingtin DNA and how our understanding of its structure can inform the development of therapies.
DNA interruptions
First up is Dr. Galen Wright from the University of Manitoba, who will be discussing how small variations in the huntingtin gene affect the course of HD. 2023 marks the 30th anniversary since the mapping of the HD gene, huntingtin. This gene is VERY big. Much bigger than most other genes in our bodies, which can make it challenging for scientists to study.
Galen is recapping what we’ve learned about the HD gene, the tendency of CAG repeats to expand in some brain cells over time (somatic instability), and the other genes that influence this expansion.
Three DNA letters code for a single amino acid, the building blocks of proteins. CAG codes for glutamine. Interestingly, CAA also codes for glutamine and it turns out that most folks with HD have a CAA “interruption” in their CAGs. People who don’t have this CAA interruption in their huntingtin gene get disease a lot earlier on in life, even though the protein coded for by the gene is exactly the same. This happens very rarely, but it suggests there is something about the DNA code which is important in HD. Scientists thought that these CAA interruptions would alter how the huntingtin gene might change through somatic instability, but it turns out that isn’t the case. This means there is still more work to do to understand what is going on.
When people undergo predictive testing, the overall length of the CAG repeats is measured. Although small DNA letter changes can make a big difference in HD symptoms, we are not at a point of measuring this in individuals to understand their likelihood of early or late onset. Interestingly, there are other diseases which are caused by DNA letter changes in the huntingtin gene, including Rett syndrome and another disease called LOMARS. These diseases also affect the central nervous system like HD.
Galen’s team mined large open datasets which bring together gene association data from many different studies, not necessarily focussed on HD. They found that the huntingtin gene is linked to traits like aging and psychological symptoms. Together, this means that the huntingtin gene is probably important in lots of different roles in our nerve cells and that the biology of huntingtin is complex. Galen rightly points out that the more we learn, the more questions we have about huntingtin.
Dissecting DNA repair
Next up is Dr. Anna Pluciennik from Thomas Jefferson University. Anna’s lab studies how mutations in our DNA letter code occur and how these might lead to disease. The mutations are caused by damage to DNA, which happens an estimated 50,000 times daily! We have evolved many ways to repair DNA to avoid a build up of mutations.
Anna’s team studies a specific type of DNA repair called mis-match repair, which corrects a situation where the 2 strands of the DNA helix aren’t properly matched so the helix structure is a bit wonky. These wonky structures are recognised by special machinery which can then try to fix these problems to correct the DNA letter code. Ironically, in some cases (like with CAG repeats) this machinery actually makes things worse.
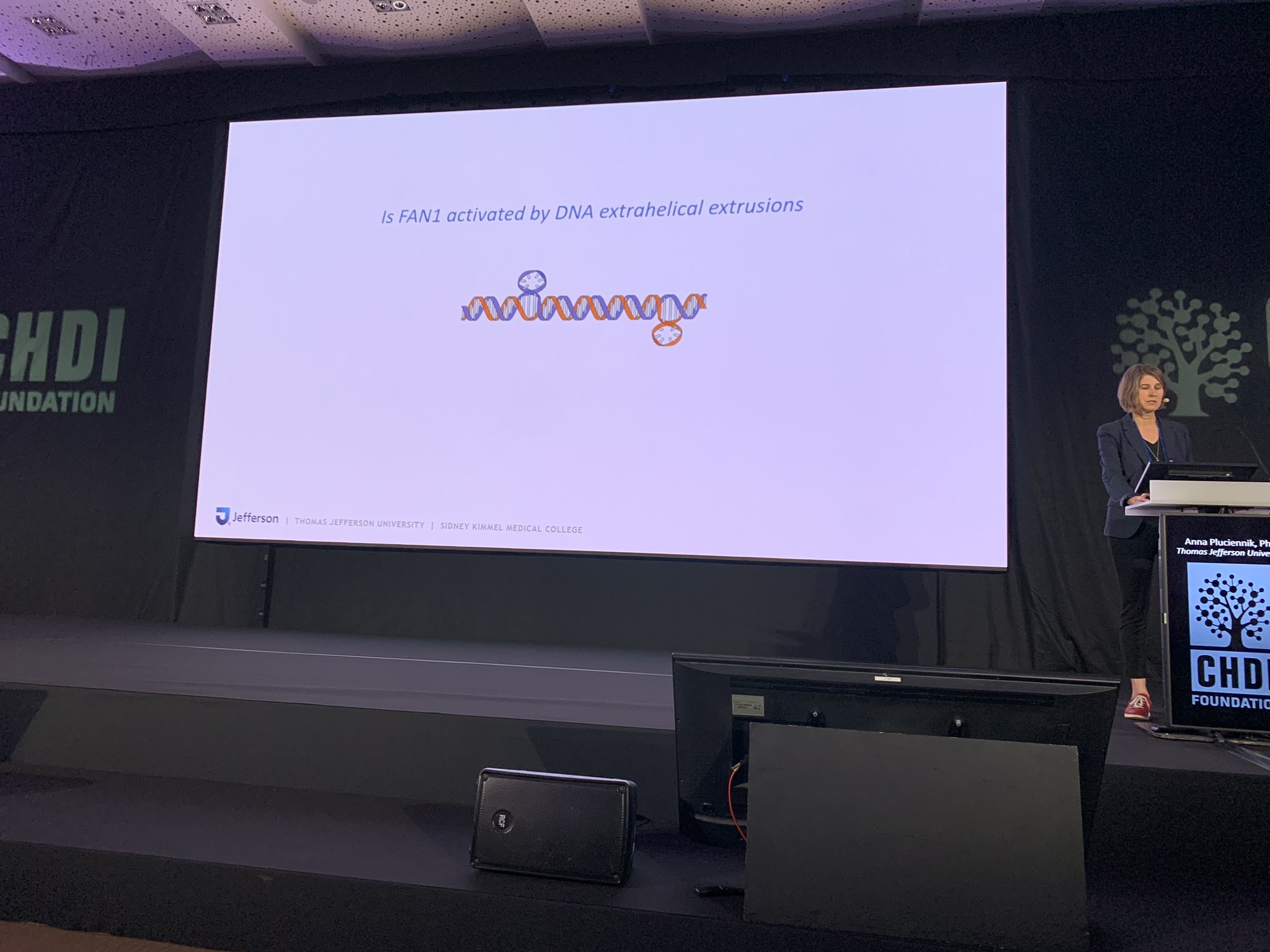
Anna’s lab studies biochemistry and she likened it to disassembling a car into its thousands of parts to understand how they all work together. This allows her team to work out details that can’t necessarily be observed in complex cell cultures or in animal models. In her lab, Anna’s team makes a proxy for the HD mutation to understand how repair machinery might recognise and try to fix it. She studies the expansion of CAG repeats, which can cause them to stick out of the DNA helix, a structure known as an “extrusion.”
Using this proxy, Anna’s lab is dissecting which DNA repair proteins do what. This type of dissection is important for future studies that might target such proteins with drugs which could help to treat people with HD. Anna’s work is helping to understand how different amounts of each protein might tip the balance to decide whether the machinery corrects DNA damage as it should, or inadvertently makes things worse.
DNA structure influences function
The final talk of this first morning session is from Dr. Natalia Gromak, from the University of Oxford. Natalia’s team studies special structures called R-loops which may be important in HD. R-loops are formed when the messenger copy of the DNA code, called RNA, is being made. If the message RNA copy interacts with the DNA like a zipper, it forms a sort of bubble in the DNA.
These structures have important roles in certain functions in cells, but can also interfere with things causing disease, so they must be balanced carefully. Very early on, a link was made between R-loops and neurodegenerative disease including ALS. Natalia’s group has generated a list of proteins that interact with R-loop structures in hopes of understanding their roles in biology and how these might go wrong, causing disease. 50+ diseases have repetitive DNA sequences which are expanded – just like in HD.
The Gromak lab found that R-loops are formed in regions with repetitive DNA, and have studied R-loops in Friedreich’s ataxia. The question for this conference of course is whether R-loops play a role in HD. Natalia’s group found that there are more R-loops in blood cells derived from HD patients, and found the same result in HD mutation-bearing neurons grown in a dish. There is also more DNA damage in both of these cell types. Next questions for the team are whether R-loops form on the repetitive sequence in the HD gene, whether they can affect the further expansion of this region (somatic instability), and whether huntingtin lowering has any effect on the R-loops seen in HD cells.
CRISPR and HD
Up next following a caffeine break is Dr. Michael Brodsky, from UMass Chan Medical School. Michael’s lab uses CRISPR technologies which can be deployed in the lab to make very precise edits of genomic DNA sequences. Targeting the root cause of HD, the CAG expansion in the huntingtin gene, is the most sensical way to treat the disease, but this is easier said than done. Gene editing would be one way to do this, but we’ve had to wait for the technology to catch up.
10 years ago, this was all a pipedream but technologies have improved so rapidly that we are now very seriously studying gene editing as a possible therapy for HD which is very exciting! Michael points out that gene editing is permanent, so much care must go into making sure there are no unintended changes. Another challenge for using gene editing for HD is that the drug must be delivered into neurons, which is no mean feat. The gene editing must also be very precise. This means that ideally only the expanded huntingtin gene is targeted, so there are limited or no changes to the normal huntingtin gene – also a tall order.
Michael’s group is taking two approaches to specifically gene edit the expanded huntingtin gene. The first is to target small letter changes (called SNPs) in the rest of the huntingtin gene DNA which tend to be associated with the expanded version. The Brodsky lab is first trying out these experiments in all sorts of different HD mouse models, results of which suggest that they are able to specifically edit just the expanded huntingtin gene – great news!
An alternative approach to specifically gene edit the expanded huntingtin gene is to actually reduce the size of the CAG expansion back to the normal range. Michael’s group has been successful doing this in HD mice and cells in a dish. There are still some kinks to work out before this can be developed as a potential treatment for HD but they are cautiously optimistic that further research will help to define a path forward.
More CRISPR and HD!
The next talk, from Dr. Ben Kleinstiver of Harvard/MGH, will also focus on DNA editing. He runs a genomic technology development group that is working on how to alter the expansion of CAG repeats and eventually create therapeutics. Ben’s lab focuses on the many ways that CRISPR can be used to make many different types of changes to DNA. They are engineering the CRISPR machinery to tailor these changes even further.
His main research question is, “what genome editing tools can be used to alter or shorten CAG repeats?” The lab takes different approaches to cutting repeats, interrupting them, or replacing single DNA letters or sequences. Because CRISPR evolved as a way for bacteria to combat virus attacks, there are still some limitations to using the CRISPR machinery to treat diseases. Ben’s group is working on overcoming these limitations to allow better access to different parts of the HD gene. Techniques include using different types of DNA-cutting or letter-replacing enzymes, and applying different methods to direct them at DNA sequences. Then they measure whether CAG repeats get shorter. The goal is to fine-tune the editing and customize it for the huntingtin gene.
This is Ben’s first HD conference! It’s exciting to see how CRISPR experts are directing their efforts towards HD. As technologies continue to advance we hope they can be applied to future human therapeutics.
Even more CRISPR!
Next up is Kathryn Woodburn from Life Edit Therapeutics who will give the last talk before we break for lunch. Kathryn works on ways to target the expanded copy of the huntingtin gene with editing technologies. Life Edit Therapeutics is looking at how different versions of the CRISPR machinery, especially those found in plants, can be used to customize editing of the expanded huntingtin gene.
Their approach to treating HD will be to use viruses to deliver their editing machinery to the brain. So far they have tried this in different kinds of HD mice with different versions and different doses of their gene editing drugs. They are able to decrease levels of harmful huntingtin protein by 40% while leaving the healthy protein intact! To get expanded huntingtin gene specificity, their approach is to target specific DNA signatures which are only found in the expanded version of the gene. Life Edit Therapeutics are looking at a few different signatures to do this and so far the data looks promising.
Making sure that there are no unwanted off-target effects is a challenging task and the scientists at Life Edit are working to get this figured out as quickly as possible. That’s all for the morning’s session!
HD Genetic Modifiers
Day One’s afternoon’s session will focus on progress being made in the study of HD genetic modifiers.
Understanding MSH3 in HD
Large scale human genetic studies, known as GWAS, have allowed researchers to identify these genetic modifiers, other genes that influence when HD symptoms begin. The first talk is from CHDI scientists, Dan Felsenfeld and Tasir Haque, who will be telling us all about their big team effort studying a gene identified in the GWAS called MSH3, and how they might be able to make drugs targeting this protein.
MSH3, as you may remember from the earlier talks, recognises wonky bits of mismatched DNA which need to be fixed. The CAG expansion in the huntingtin gene is prone to create these wonky bits, and it’s thought that MSH3 activity at the huntingtin gene may inadvertently increase the number of CAG repeats in brain cells (somatic instability). Scientists think that MSH3 could be a good target for medicines as switching off this gene seems to be beneficial in animal models of HD, as it reduces somatic instability – the expansion of the CAG number – in the huntingtin gene.
Completely switching off a gene is quite challenging to do in people, so instead scientists are making so-called “small molecule” drugs, which could potentially be taken by mouth, that aim to stop MSH3 working so well in cells. Dan’s team has considered different ways to inhibit MSH3 and created a toolbox of materials and protocols to study their small molecules. This will help other researchers hoping to make drugs targeting MSH3. To make better drugs, it helps to be able to “see” the MSH3 protein. Using clever techniques, it is possible to create 3D models of the protein, and then the scientists can see where and how their molecule binds.
Tasir Haque is now showing some animations that zoom in on different parts of the MSH3 protein and where the drugs fit in. Lots of squiggles that have great meaning for structural biologists! Using these models, they can work out how to better improve these early stage drug molecules to better fit into all the nooks and crannies of the MSH3 protein surface, which should improve their properties.
Drugs targeting MSH3
Next up is Caroline Benn from LoQus23 Therapeutics, a company that is also working to develop drugs targeting MSH3 – it’s a hot area! LoQus23 is taking a slightly different approach to the CHDI program on MSH3 – they are making molecules which target a different region of the protein. This is good news for the HD field as it’s great to be able to test out multiple approaches! Although their approach of targeting different regions of MSH3 is more difficult, they have pulled it off and found two such series of molecules which are very potent and selective, meaning they bind the MSH3 protein very tightly without affecting other proteins.
LoQus23 has also established a way to measure somatic instability in cells in a dish to test how well their molecules work. These are complex experiments which take weeks from start to finish.They will also be able to use this platform to find new targets, besides MSH3, which play a similar role in this part of DNA damage repair which is so important in HD.
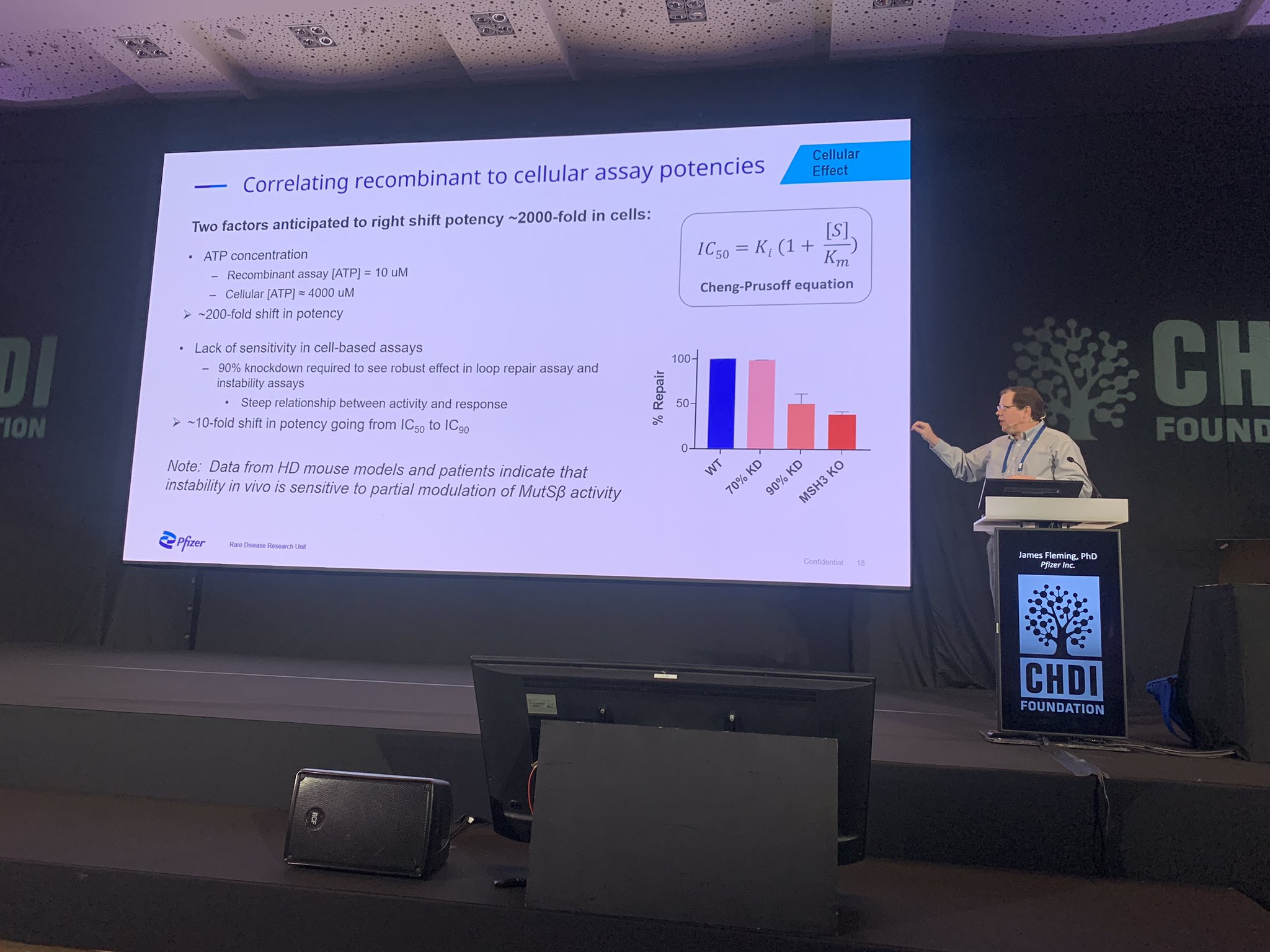
Next up is James Fleming from Pfizer. This company is also developing drugs to target the pathway involving MSH3. Pfizer are taking a similar approach to the CHDI team, and like the other folks, have developed a suite of tools and methods to test their molecules for the ability to stop expansion of CAG repeats.
Like other pharmaceutical companies, Pfizer takes a series of steps to screen potential drug compounds, understand how they interact with the proteins they target, and then test them in cells and in animals. They too are using 3D models and chemical tests to show that their drugs can stick to the protein complex that MSH3 is a part of, which has helped them make these molecules better and better over time. The next step is to test these drugs in cells grown in a dish. A lot of this work is focused on the minute details of the protein chemistry, structure, and energetics. Suffice it to say that mathematics figures into the drug development process!
To bring these studies into animals and then later into humans will require a drug with the right properties: the ability to target MSH3, the ability for the body to break it down, and the ability for the drug to get into the brain. Not a small task! After better understanding the properties of a new drug in cells and animals, it may then be tested for safety in people. Right now this is a bit far in the future for all of the compounds presented today, but it’s exciting to see that companies are moving the work forward.
Massive datasets to identify genetic modifiers
Kicking off the last session of the day is Jim Gusella from Harvard, who will be telling us about genetic modifiers of HD on behalf of a large consortium of scientists who study HD genetics.
Jim begins his talk by acknowledging all of the HD families who have so generously shared their data and samples with HD researchers over the years, without whom, these large-scale analyses would not be possible. An interesting finding we have known about for a while now, is that folks with the same CAG number may start to have symptoms at very different ages. Genetic modifiers are markers in the DNA which can explain this early or late onset of symptoms.
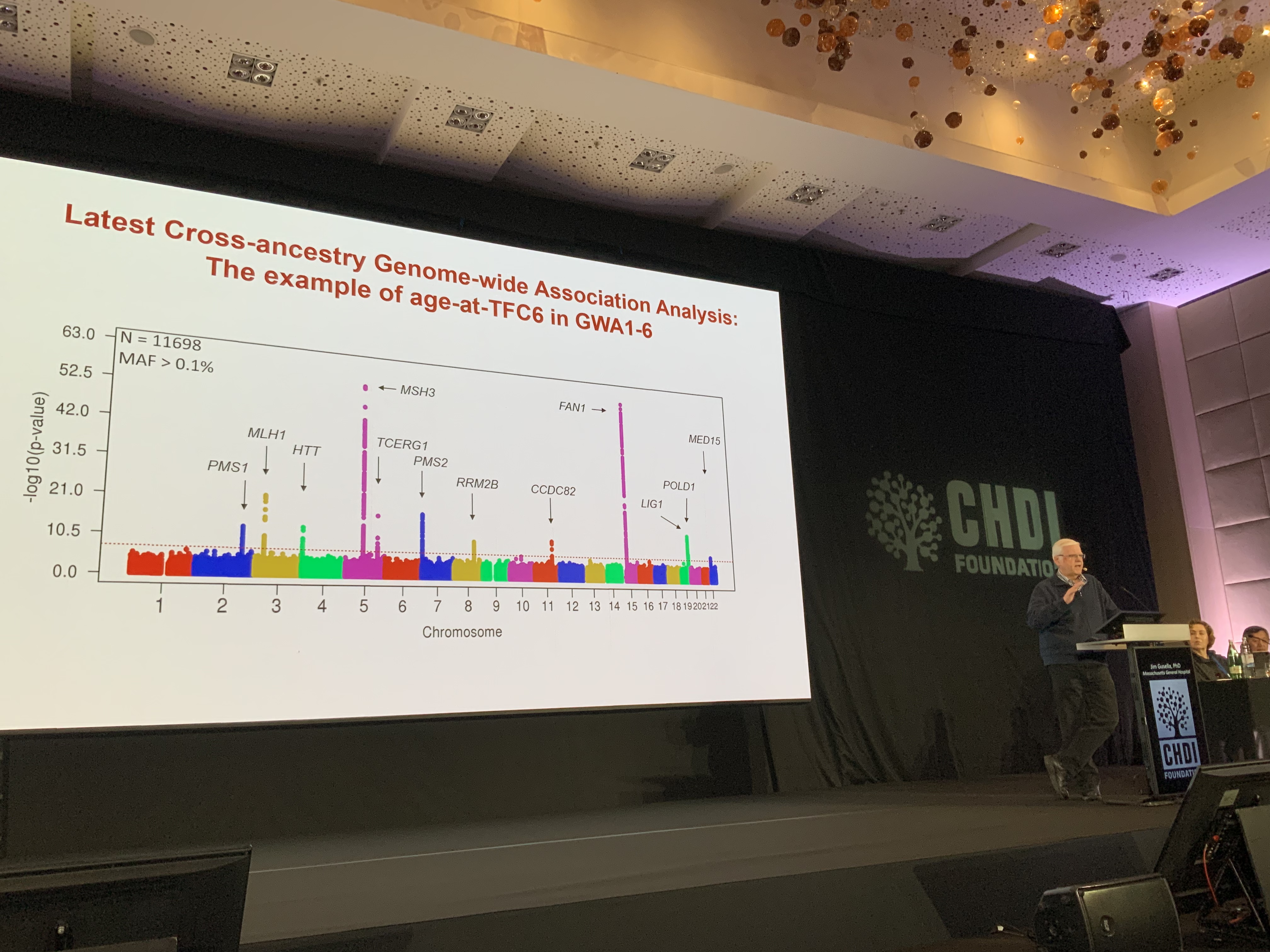
More and more evidence is pointing to a particular driver of the onset and speed at which HD gets worse over time: the expansion of CAG repeats in some cells. This process, known as somatic instability, seems to be linked directly to genetic modifiers. The power of these modifier studies comes from the number of patient samples which are analysed – more data means higher confidence conclusions. In the most recent study, 11698 participants’ data were analysed which is amazing!
A problem with HD research, and science in general, is that many of the samples analysed are from Europeans or folks of European heritage. In this later dataset, the team is working to include a more diverse group of patients in the data. With such a wealth and diversity of data, it is possible to zoom out on a large scale and make general predictions of how genetic modifiers – tiny changes in other genes – affect when people with HD might reach certain stages of HD.
It is important to emphasize that this is a way to have more confidence about what other genes most affect HD in people. This is different from being able to predict onset or disease course in an individual person with HD. Jim’s team and the genetic consortium are also looking at how subtle differences and “interruptions” in the sequence of CAGs in the huntingtin gene affect the DNA structure and the tendency of the repeats to become unstable and grow longer. The good news from this most recent dataset is that MSH3, the topic of the previous session, is still a very significant modifier by all of the analyses Jim and colleagues used. That lends a lot of credibility to all of these approaches targeting somatic instability and trying to stop the expansion of the CAG repeat or shrink it.
CAG expansions in specific brain cells
Next up is Nathaniel Heintz, based at Rockefeller University, who will be talking to us about his work on understanding which genes are switched on or off in HD. The Heintz lab developed a series of techniques that allow scientists to “sort” the nuclei of many different cells and look at genetic messages in many cell types. This has become an important way to study why certain cells are most vulnerable in HD and other diseases. These analyses use post-mortem brain tissue samples, enabled by the amazing generosity of the HD community.
The striatum, an area in the center of the brain, is most deeply affected by HD. Heintz’s team is able to sort through different kinds of cells in the striatum and have discovered that CAG repeat expansion occurs most often in one type of cell, medium spiny neurons.
We have known for a long time that medium spiny neurons (MSNs) are lost in great numbers in HD. There are different types of MSNs and oddly it turns out that both those which are vulnerable in HD and those which survive are subject to CAG repeat expansion. The reason these medium spiny neurons seem to have higher levels of expansion could be due to the higher levels of MSH3 which are found in these cells but that link is not yet proved. Medium spiny neurons also have huge numbers of genes that are switched on or off in HD – 1000 turned on and 500 turned off! Many of the genes affected are involved in DNA damage repair – again reinforcing the important role this likely plays in HD.
Ongoing work is addressing the questions of when medium spiny neurons are affected in HD and how best to intervene. They are also looking at other brain areas and going layer by layer to understand exactly which types of cells are becoming damaged or lost.
Speed of CAG expansions
The last speaker for the day is Steve McCarroll, from Harvard. Steve’s lab looks at which genes are switched on or off at the levels of single cells, instead of a big mixture of loads of different types of cells – an incredibly detailed approach. He uses a fruit analogy to talk about the power of this technique – you can compare cell types like different kinds of berries, the same cell type in different people like apples to apples, the differences between different cells of the same type, like looking at two blueberries.
From these single cell analyses, they can work out which cells disappear over the course of HD, confirming previous findings that medium spiny neurons and spiny projection neurons are the most vulnerable kinds of nerve cells. They can also work out exactly which cells have CAG expansions – this seems to suggest that the vulnerable medium spiny neurons are the most likely to have the most expansion. The CAG expansion during a person’s lifetime in these cells seems to be very specific JUST to the huntingtin gene, and only the HD gene, not other genes which have similar kinds of DNA code. The majority of these vulnerable brain cells have moderate expansion in the CAG repeat, but a small subset have huge expansions which scientists haven’t quite figured out the reason for just yet.
According to the McCarroll lab’s data, the moderate expansions seem to happen very slowly over time, but the more exaggerated expansions happen much more rapidly. The key question is, at what threshold of CAG repeat number does the expansion speed up, causing damage and death of these vulnerable nerve cells? To try and figure this out, the McCarroll lab can compare individual neurons with different CAG numbers, and split them into groups to better understand what lengths are most problematic. They grouped cells by CAG number and strangely didn’t see too many differences in genes turning on and off at lower repeat lengths. The most profound changes occur in cells with very, very long CAG repeats, more than 180.
McCarroll is proposing a very different way of thinking about HD pathology and how the disease works over time. There is some interesting chatter in the audience! But this is why it’s so good that all these scientists can get together at this meeting to debate all these ideas.
Tune in tomorrow!
That’s all for today, folks. We’re breaking for the night, but will be back tomorrow morning! Remember to continue to follow live updates for the rest of the conference with the hashtag #HDTC2023.
For more information about our disclosure policy see our FAQ…


