
Huntington's Disease Therapeutics Conference 2025 – Day 3
HDBuzz reported live updates on Bluesky from the 2025 HD Therapeutics Conference. Read on for coverage of Day 3. #CHDI2025


We’re back for the 3rd and final day of CHDI’s Huntington’s Disease Therapeutics Conference!
New Technologies and Breakthrough Science
This morning’s talks are kicking off with new technologies that have the potential of leading to breakthroughs in science for neurodegenerative diseases, like HD, and transform the field.
Ross Wilson: Molecular Scissors To Chop Out CAGs
Our first speaker is Ross Wilson, who is working on using CRISPR to selectively remove the disease-causing CAG repeats within the huntingtin gene. Whoa!
CRISPR is a molecular technique that began taking the scientific world by storm when it was broadly introduced in 2012. Just last year the first CRISPR-based drug was approved for blood diseases, like sickle cell anaemia.
You can think of CRISPR like molecular scissors. Researchers can use these molecular scissors to very precisely edit the genetic code to make all types of different changes. It’s a super powerful technique with many applications for diseases, including HD.
Ross recaps work by others who showed that CRISPR editing of the HTT gene increases lifespan and improves behavioural characteristics of HD mouse modes. However, this approach targeted both copies of HTT – totally shutting off both copies of HTT wouldn’t be a good therapeutic.
An updated approach he presented uses some genetic tricks to use CRISPR to only target the disease-causing copy of HTT. This was a great proof-of-concept paper, but there were concerns with “off target” effects – unintended editing of other genes that could come from this approach.
Ross’ lab has combined CRISPR strategies for improved efficiency and precision – selectively hitting just the expanded HTT. His big improvement is to break down CRISPR machinery after it does its job. If it sticks around in the cell, it can increase the chance of off target effects.
Ross went into some technical details about exactly how they’re getting the CRISPR machinery to shut off after it edits expanded HTT. The trick seems to be to deliver CRISPR pre-assembled. This is something that has only been possible because of recent achievements.
Ross highlighted some advantages of his system: it’s easy to manufacture, he can scale production quickly, it’s smaller than other approaches (which matters when you want to get things into the brain!), AND it turns off after it does its editing job. Quite a long list of advantages!
“Within a genome of 2 meters of DNA per cell that codes for over 20,000 proteins, CRISPR base editing is like being able to target a specific letter in a single word from a library of hundreds of books and know that is the single letter that you want to change. The specificity is AMAZING!”
While it may be possible to deliver this into the brain using a harmless virus, Ross is describing an alternate method that uses special types of small particles that can carry the CRISPR ingredients into the brain in a ready-to-go format.
His team is also working on improving ways these types of drugs can be delivered to the brain, which currently require brain surgery. They’re testing various approaches in mice, but the hope is that they’ll be able to adapt their findings to people one day soon.
When they tested the effects that their potential drugs have in mice that model HD, they showed they can specifically edit just the expanded copy of HTT using CRISPR. Ross highlights that this competes with the currently best-in-class CRISPR approaches being used. Great news!
They also tested their CRISPR tools in pigs to get an idea of how well it works in larger animal models with a brain closer in size to that of people. No one likes to experiment on animals, but this is the best way we currently have to get drugs tested for safety before clinical trials in people.
In the pigs, they found some limitations with their approach around how well the material was distributed throughout the brain. These experiments are helping them improve their approach. They already have some ideas of how to improve the system, such as a “shell” for the CRISPR machinery, decorated with keys that fit into the locks of certain cells (neurons!). This is a HUGE advantage since we know that HD preferentially impacts certain cells in the brain.
Their updated approach looked to improve things in pigs, so with that improved strategy they’re going back to mice that model HD to see how their new and improved CRISPR machinery may influence signs and symptoms of HD.
Despite the advantages of the CRISPR delivery particle, it may be easier to move to the clinic using the harmless virus approach, so Ross’s group is exploring that too so they can get to the clinic as fast as possible. Exactly what all HD families and researchers want!
Zaneta Matuszek: Editing Single Letters In The Genetic Code
Our next speaker is Zaneta Matuszek, who recently graduated from the lab of David Liu at MIT. She’ll be sharing a similar story about precisely editing the CAG repeats, but with the goal of reducing somatic instability.

While we know that the length of the repeat within the HTT gene strongly contributes to the age of symptom onset, we also know there are other factors at play. The exact code of the repeating DNA seems to really matter. Small interruptions in the CAGs seem to have a big influence.
Zaneta is trying to capitalise on these small changes that can have a big impact. She uses a super cool variation of CRISPR called “base editing”. This is an ultra specific version of CRISPR that lets her edit single letters within the genetic code.
Within a genome of 2 meters of DNA per cell that codes for over 20,000 proteins, CRISPR base editing is like being able to target a specific letter in a single word from a library of hundreds of books and know that is the single letter that you want to change. The specificity is AMAZING!
Zaneta is using base editing to alter the CAG repeats with interruptions of CAA – something we know can delay the presence of HD symptoms in people by up to 13 years. She has data to show that she’s effectively able to do this in cells grown in a dish.
She’s also looked at how this may influence somatic instability. Excitingly, her data shows that the CAG repeat tract doesn’t get bigger when she uses base editing to alter some of the CAGs to CAAs in the cell model they are using for this in the lab. Zaneta thinks that this suggests that this approach could be a good way to go about controlling somatic instability, but also that CAA interruptions themselves might be influencing somatic instability in some way.
They also looked at other genes with long runs of CAGs to see if there are off target effects in those genes. While they mostly saw changes in the HTT gene, there were also changes in these other CAG-containing genes. So there is some work to do to make sure this approach is safe and on target.
Next they moved into mice that model HD. In these mice, they were able to target the expanded copy of HTT and also have data to suggest somatic instability is more stable. They also seem to see repeat contractions! Zaneta highlighted that the team are busy in the lab looking into this more.
William McEwan: Hijacking The Cell’s Garbage Disposal
Up next is William McEwan, who is working to target a protein called TRIM21 that he hopes will have a positive impact on both protein clumping and somatic instability in HD. William begins by talking about neurodegenerative diseases more broadly, starting the discussion with Alzheimer’s and a protein called Tau that causes protein clumping in that disease. In Alzheimer’s, Tau clumps can spread from cell to cell in the brain, something called “seeding”.
“We’re living in the age of artificial intelligence (AI)! And it’s exciting to see these new tools being applied to HD research.”
William highlighted that we have the ability to remove these Tau clumps from external compartments within the cell, but not the major internal compartment, called the nucleus. It’s protein clumps here that seem to be responsible for causing disease features of Alzheimer’s.
His work focuses on a protein called TRIM21. TRIM21 is a receptor – a protein stuck out on the cell surface like an antenna to catch cell signals. The signals TRIM21 catches are antibodies, specialized immune proteins that keep us healthy and help with fighting disease. William’s work suggests that TRIM21 could allow for the breakdown of Tau within the cell. Learning more about this interaction could help advance therapeutics for Alzherimer’s and help us learn about similar mechanisms that exist across neurodegenerative diseases.
William is exploring TRIM21 in HD by looking at how it may interact with HTT protein clumps in cells grown in a dish. One of his goals is to add special molecular decorations to help degrade these clumps. It’s always great to see what we can learn from other disease fields and new tools to apply to HD.
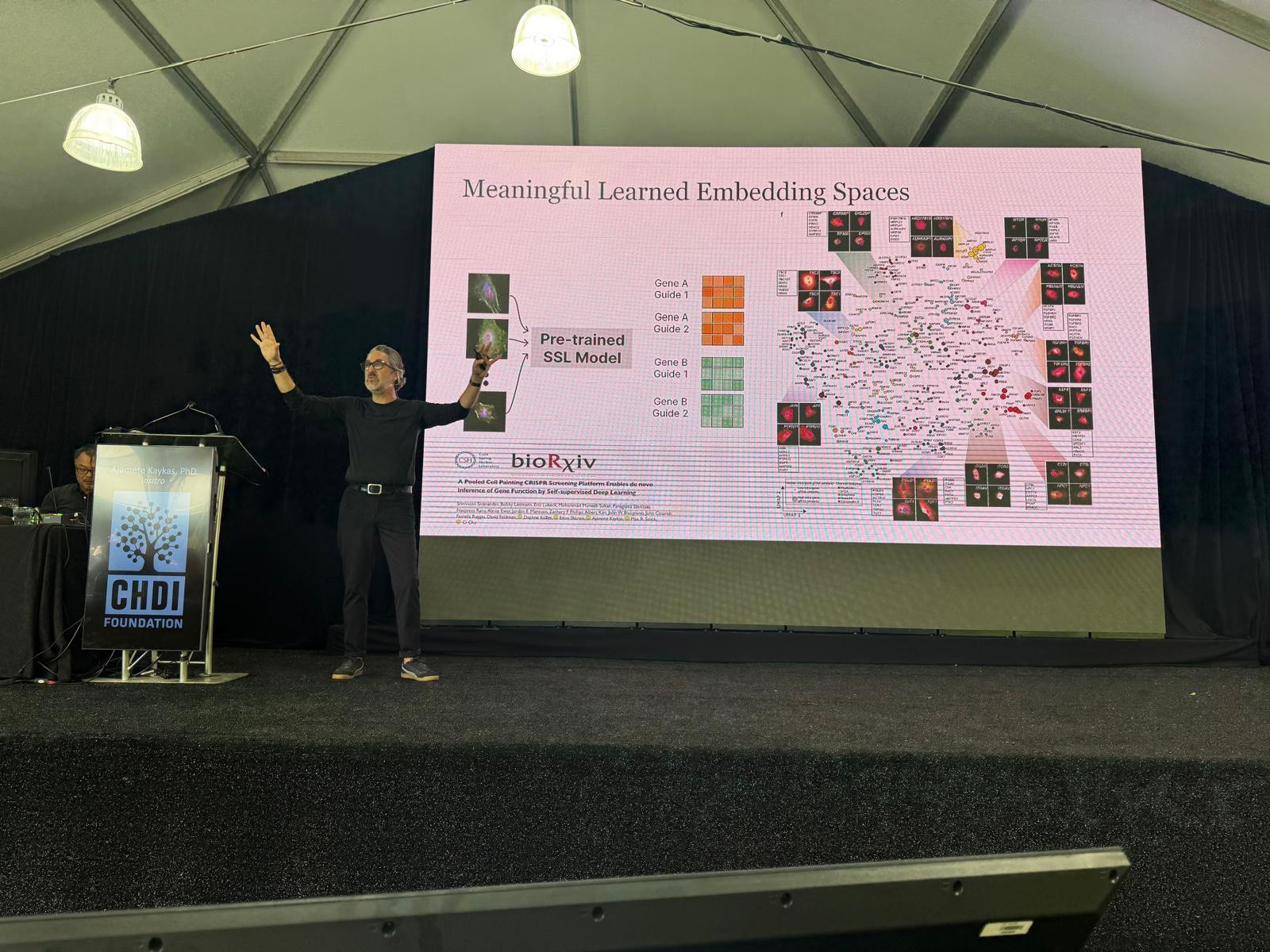
Ajamete Kaykas: AI-Guided Insights Into HD Drug Discovery
Our next speaker is Ajamete Kaykas from Insitro, a company that’s trying to use computers and machine learning to help us understand disease better and drive clinical decisions for therapeutic development with AI-guided insights. His focus today will be on their biological discovery platform that they’ve focused on human genetics for drug discovery to identify targets that they can advance. We’re living in the age of artificial intelligence (AI)! And it’s exciting to see these new tools being applied to HD research.
After they use their computer wizardry to identify new possible drug targets, they then move into cells grown in a dish to validate their findings – computer predictions alone mean very little without real world validation. They’ve scaled other technology platforms developed by others that allows them to quickly visually scan cells using fancy robotics in a completely automated way. This is technological advancements at their best to save hard working scientists lots of time and effort!
There are lots of advantages to using this platform, the first of which is that it’s great for looking at neurons! Neurons are a sensitive cell type that don’t like to be lifted off of cell dishes once they’re there. But that’s actually what a lot of traditional experiment approaches require. Insitro’s method allows the neurons to remain in the dish while they’re analyzed. This preserves information that is lost with other techniques. Neurons are shaped like a tree, with a trunk, body, and branches. Preserving these structures for analysis can be very informative for diseases like HD.
He shared that they’re using this to look at TDP43 – a gene we mentioned yesterday for its involvement in ALS. Using their system to analyze cells affected by TDP43, they pull out biological mechanisms already published, helping them to validate the approach they’re using. With enough data like this they can train their machine learning system to learn about the cells in great detail. Excitingly, they’re hoping to use this technology to start predicting biology that’s influenced by disease, without even doing experiments. Every PhD student’s dream!
So far, they’ve found that their machine learning models outperform other, more old-fashioned analyses. They can even use their models to predict where TDP43 would be located in cells to help predict disease. Incredibly cool! Briefly, he shared what he thinks they could do for HD. They could set up computer experiments to help identify new connections in human genetics and biological pathways to help us advance drug discovery. These technological advancements are an exciting way to speed our way to an HD treatment.

Kathleen McGinness: Getting To Undruggable Targets Through RNA
Next up is Kathleen McGinness from Arrakis Therapeutics. Her team is working to develop small molecule drugs which target RNA message molecules. RNAs are the message molecules which contain the instructions for making different protein molecules in the cell.
Traditional drug discovery has targeted proteins but now many companies are going after the RNA molecules insteads. That’s because many proteins aren’t druggable or are very hard to target with small molecules so instead, targeting the RNAs that encode them gives us another way to hit these targets with drugs.
Arrakis have a whole suite of tools to pursue small molecules for a given RNA target. They are trying to find small molecules which hit many different RNA message molecules. These small molecules come in lots of flavours and might have different effects on the RNA molecule. This includes blocking interaction between how RNA and proteins stick together, changing how RNA molecules are edited into their mature functional forms, as well as stopping production of the protein they encode.
One of the lead programs at Arrakis is focussed on Myotonic Dystrophy (DM1), another repeat expansion disease. For DM1, the RNA message molecule itself is thought to be what causes disease signs and symptoms. This message molecule contains a long string of toxic CUG repeats.
Arrakis have developed small molecules that bind the CUG repeat which they have tested in the test tube, cells in a dish, and animal models. These molecules block a protein called MBNL1 from binding onto the RNA message and the team has precisely determined how they bind to the RNA.
They also found that these drug candidates seem to help recover some of the signs of DM1 in cells and mice back to baseline at a molecular level. This included how gene messages are processed, protein clumps in the cell, and muscle symptoms in mouse models of DM1. Arrakis are at this conference because they’re thinking about applying their technology to HD, and going after drug targets which have proved challenging with more traditional approaches so far. Woohoo! More people focused on HD!
Andreas Mund: Mapping Protein Levels On Top Of Brain Samples
Next up is Andreas Mund, who is using a super cool method to look at where different proteins are within a tissue sample. This is a massive advancement from standard techniques that smush up samples and look at bulk protein in a tube but give no information on where things are in tissues or cells.
“With enough data like this they can train their machine learning system to learn about the cells in great detail. Excitingly, they’re hoping to use this technology to start predicting biology that’s influenced by disease, without even doing experiments. Every PhD student’s dream!”
These new techniques takes slices of tissue, like brain slices, puts them on microscope slides, then analyzes them with different probes against various proteins to reveal where proteins are within the tissue sample. They then put all the data together to build a big map of protein levels on top of the tissue. They can zoom in on the tissue samples and look at single cells that make up the tissue, where they can figure out where 1000s of proteins are located and who they are hanging out with. This super detailed analysis can give all sorts of insights about biology in health and disease.
Andreas is showing us some data where they used this technology to study an autoimmune-related skin disease. Using techniques that analyzed bulk protein in a tube, they could see that certain proteins involved in the immune response were elevated. Using their cool spatial protein technology, they looked to see exactly which type of immune cells had these kinds of changes and where they were in the layers of the skin. It turns out there is a drug that hits the elevated protein, so they treated a model of this disease which prevented onset of symptoms. Amazing!
Next, they showed this treatment also worked in people suffering from this skin condition after all drugs previously failed. A great success story! They are seeking to expand this platform to work in other disease areas, HD included, and use machine learning to provide new insights to their data that people might miss.
Looking at samples from the brain, they see that location matters: the same cell type in different areas of the brain have different proteins arranged in different ways. They are just getting started on HD, so we look forward to seeing their findings soon.
Clinical Biomarkers In HD Research
We’re back for the last session of the conference! And they saved a good one for the end. In this session we’ll be hearing about clinical biomarkers in HD research. These types of biomarkers will help clinicians eventually determine things like when HD symptoms begin and how rapidly disease will progress.
Jim Rosinski: Applying Machine Learning To Biomarker Identification
Our first speaker of this session is Jim Rosinksi from CHDI. Jim is a self-proclaimed data geek and is applying protein analysis and machine learning to the HD-Clarity dataset to identify biomarkers to help predict disease stage and progression.
HD-Clarity is a study that collects spinal fluid and blood from people with HD, led by HDBuzz superstar emeritus, Ed Wild. These samples help us find new biomarkers of HD so we can track disease progression, and figure out how well possible treatments might be working.

Jim is explaining how amazing the data are because of how many people from the HD community have generously participated. He calls it “ridiculously high quality”. They have so much data now, that he can’t tell us about it all in one talk! So he will focus on the proteins found in the spinal fluid.
Once the samples are collected from sites around the world, they are analysed by specialised scientists who figure out what markers are present and how they might track with HD. Encouragingly, all the expected biomarkers come up as strong hits. This is important because it validates what we already know and lends credibility to the scientific approach being used. This includes some biomarkers HDBuzz readers will probably know, like huntingtin and NfL, as well as newer ones, like NPPB.
One of the new ones Jim and his team have been looking at recently is NPPB, or natriuretic peptide B, which goes down as HD progresses. This is an established biomarker for heart disease where high levels are bad. So NPPB going down means the marker is trending in a positive direction for people with HD.
Jim comments that CSF biomarkers seem to be much simpler to figure out and identify than those in blood. We probably need more samples and more complicated analyses to figure out blood biomarkers completely.
The next step for biomarkers is to use them to predict disease stage. Jim and other biomarker data nerds have been collating tons and tons of data from people with HD as their disease progresses to feed machine learning programs to see what patterns they can spot and use to make predictions.
There are all types of questions they can ask with these models. If they have CSF data from 2 people, can the program correctly identify who has HD? Where are they in disease progression? Turns out that the models are pretty good at this! They can figure out pretty well who doesn’t have the gene for HD and at what stage of disease they are. It will take a little more work to be able to distinguish between early stages, where outward symptoms are not apparent. For example, distinguishing HD-ISS stage 0 and 1 is very challenging for these models so more work is needed there.
Rather than relying on looking at the levels of just one protein, like those we already know about (ie. NfL), they look at the levels of a panel of proteins. This seems to be critical for making accurate predictions with this model. The good news is that once other types of molecules are added to the protein analysis, the model should get better.
Next, they are looking to expand these models to other types of biofluids, looking at other types of molecules in these samples, like fats, and building new models for these types of predictions. These models will be especially helpful for understanding the impact of different therapeutics in clinical trials.
Leslie Thompson: Cell Free DNA As A Biomarker
“These new techniques take slices of tissue, like brain slices, put them on microscope slides, then analyze them with different probes against various proteins to reveal where proteins are within the tissue sample. They then put all the data together to build a big map of protein levels on top of the tissue.”
Up next is Leslie Thompson from the University of California, Irvine. She will be telling us about her research on cell free DNA and special chemical decorations on DNA which could be used as a biomarker for HD. This type of approach was originally proposed as a biomarker for cancer research. The hope is to transfer those successful techniques over to HD.
Leslie is using Enroll-HD and Clarity-HD samples. So another big thanks to all who have participated in those observational studies!
But what exactly is cell free DNA? The idea is that people take a blood sample, which contains free-floating, fragmented pieces of DNA. Thus, the DNA is free from a cell – cell free DNA. In this cell free DNA, Leslie and her team are looking at small chemical marks that decorate the DNA, one of which is called methylation, and how that might differ in people with and without the gene for HD.
There’s already a kit for liver cancer on the market in China that can distinguish between different stages of liver cancer using this technology. Cell free DNA is also being investigated for tracking other diseases, like diabetes and prenatal diagnosis.
For brain diseases, it’s being used for multiple sclerosis and Alzheimer’s. This type of testing is very sensitive, and would be a fantastic biomarker to have for HD if it does in fact track with disease progression – and that’s exactly what Leslie is trying to figure out. So far Leslie and her team have collected pilot data from a small group of people without HD or with varying stages of HD showing that they have this experimental setup working in their lab. Always the first step!
While this data is preliminary and from a very small number of people, it seems to show that there are some differences between people with and without the gene for HD with additional separation of samples based on HD disease stage. Overall, she identified lots of different targets with methylation changes and showed data from a few key genes. These types of experiments give researchers large datasets to dig into!
Leslie explained that the changes identified are primarily from blood cells, not brain cells, which only made up about 3% of the cell free DNA. Interestingly, there are some changes they identified that correlate with disease progression, supporting the potential of cell free DNA as an HD biomarker.
Excitingly, Leslie thinks this type of analysis could be used for predictions. Meaning, once they work out all the kinks for this experiment, if they were to get a sample, they may be able to predict exactly where in disease trajectory someone is to monitor disease progression.
Their next steps are to expand this study to blood samples from many more people and look at cell free DNA changes in CSF samples, which will clue Leslie’s team into what’s happening in the brain.

Manuela Moretto: Brain Scans To Find Biomarkers Of Brain Health
Next we’re hearing from Manuela Moretto from the University of Padova and King’s College London. Manuela will be talking about her research on using brain scans to track HD in the iMarkHD study.
iMarkHD is a 5 year study taking lots of measurements, including different types of brain scans and biofluid samples from people at different stages of HD, some from people who are many years before predicted symptom onset. Studies like this can help us better understand the brain changes which happen during HD in much more detail, as the same people are measured again and again over the 5 year timeframe.
They are relying on several different high tech ways of imaging the brain to paint a more thorough picture, which should help figure out some of the very subtle changes which happen in HD throughout the disease course. One of these approaches uses special tracer molecules that light up the brain when they stick on to certain markers. Many of these markers are known indicators of brain health. Seeing how much the brain lights up in different people at different stages of HD could help us understand changes to brain health in HD.
This study has done a ton of work! The findings so far are very interesting. Some of these measures showed clear differences between the groups assessed whereas others were less obvious. This helps HD researchers know the brain features to which they need to pay attention.
Manuela finishes by thanking the participants in this study, who went through so many brain scans! It is a huge commitment of time and energy, and a truly impactful contribution.
Peter McColgan: Roche Moving Toward Selective HTT Lowering
For the penultimate talk of the day, we are hearing from Peter McColgan from Roche. If you are having deja vu, don’t panic, Peter did already give a talk on Day 1 and is back on the roster for Day 3. He will tell us about Roche’s selective approach for lowering the expanded copy of huntingtin, which they are developing. This differs from the total HTT lowering approach they’re currently exploring with their tominersen trial.
The new strategy Roche is using is to target a short region of the huntingtin gene that is only found in the expanded copy of the gene. The unique genetic area they have chosen is found in about 40% of people with HD. While this obviously means that, if successful, this specific drug wouldn’t work for 60% of people with HD, if this is successful Roche would certainly work to develop other iterations that do work for everyone with HD. This is very similar to what Wave Life Sciences are doing.
“Manuela finishes by thanking the participants in this study, who went through so many brain scans! It is a huge commitment of time and energy, and a truly impactful contribution.”
They have already tested this new drug candidate in mouse models of HD and shown that their drug preserves regular huntingtin whilst lowering the expanded huntingtin. Great news!
With the knowledge that this approach seems to work in mice, they turned back to their data from people from the GENERATION-HD1 trial. This lets them see exactly how many people have the unique genetic area on their expanded HTT gene that they want to target.
One of their goals now is to identify the number of people across the globe that have this unique genetic signature and so might benefit from this new drug candidate. They’re also collecting geographic data to see exactly where in the world these people are.
During the Q&A portion, someone asked Peter when they think they’ll be ready for clinical trials for this approach that specifically targets the expanded copy. Peter said they’re planning to start trials for this approach by the end of this year. Stay tuned!
Hilary Wilkinson: Diagnosis, Prognosis, Safety – Teasing Apart Biomarkers
The final talk of the conference is from Hilary Wilkinson from CHDI. She will be telling us about different types of CSF and blood-based biomarkers that are used in HD clinical research. Biomarkers can be used for LOTS of different things in HD research. These uses could be diagnosis, prognosis, safety or efficacy of drugs, as well as disease progression.
Hillary and the team are working to answer various questions about how we can most logically apply biomarkers across all of the uses they have. Just some of those questions are, how should we collect samples to optimize detection? How do we decide on the context of use for a biomarker? What sorts of statistical tests should we use for various approaches?
They’re not just asking these questions about new biomarkers, but also old biomarkers. Some of our oldest and currently most reliable biomarkers in HD research are expanded HTT and NfL.
Hillary is sharing an experimental plan for looking at levels of biomarkers using samples from the HD-Clarity dataset. When they look at expanded HTT in CSF, the fluid that bathes the brain, they see good reproducibility across site visits. This is critical for a good biomarker!
She thinks that blood levels of expanded HTT will make a good biomarker for intervention trials because distribution levels correlate with disease severity. This helps researchers stratify people with HD by disease stage just by looking at levels of expanded HTT that they have in their blood. However, a very important caveat is that expanded HTT levels don’t correlate with HD signs and symptoms. So there are clearly limitations with using this specific biomarker for measuring certain clinical changes associated with HD.
Now Hillary is moving on to the data she has looking at NfL as a biomarker. Levels of NfL in CSF seem to tally with the amount of neuronal damage caused by HD. Also, like expanded HTT, NfL levels in CSF are reproducible across visits for people in the HD-Clarity trial. Like others have shown, Hillary also sees an increase in NfL in CSF through HD disease stages with elevated levels seen very early, before symptoms are obvious.
In looking at NfL data from both HD-Clarity and Enroll-HD, Hillary feels it could be used for selection of clinical trial participants and may be useful for clinical trial design. Having such a powerful biomarker with multiple uses would be a big advantage.
Now she’s switching gears to newer biomarkers in the HD space, PENK and PDYN, which may have potential as disease activity biomarkers. Right now they’re working on figuring out how they can robustly measure these two new biomarkers.
Hillary also brought up some of the advantages of using somatic instability as a biomarker, but there are limitations here too. We can’t measure expansions in the brain while people are alive and what we can measure in blood is very subtle. There is work to do before we can use this in clinical trials.
She ended by stressing that sample collection and storage is critical for successful evaluation of biomarkers. This remains a high priority for CHDI as well as all researchers working on biomarker development.
That’s all for this year folks! We hope you enjoyed following along and learning about all of the super cool new HD research going on all over the world.
For more information about our disclosure policy see our FAQ…


