
Re-routing the huntingtin protein inside cells
Canadian team decodes the tags that re-route the huntingtin protein within cells, helping explain how it becomes toxi

The mutant huntingtin protein causes Huntington’s disease, but not all mutant huntingtin is equally toxic. Exactly where it’s found inside cells has a major impact on whether cells can deal with the mutant protein. New research is helping understand the signals that move mutant huntingtin around the cell, provides clues about how to make it less toxic.
Huntingtin – and its evil twin
Though Huntington’s disease is due to a genetic abnormality, we know that the damage it causes to cells is due to the mutant huntingtin protein. The HD gene is a set of instructions for making the huntingtin protein. In people with HD, and those who are destined to develop the disease, a ‘spelling mistake’, at the start of the genetic instructions, causes a particular bit of the protein to be longer than normal. Too many building blocks called ‘glutamine’ are stuck together at one end of the protein.

Having too many glutamine blocks at the start of the protein has disastrous effects, and is what’s to blame for the harmful effects of mutant huntingtin.
One of the main jobs of Huntington’s disease researchers is to figure out exactly how the mutant protein is different from normal, and how those differences cause it to damage cells. Every new bit of information about the ways in which the protein causes damage, is a possible lead in the search for treatments to slow the disease down.
Ray Truant is a Huntington’s disease researcher at McMaster University in Ontario, Canada. Truant and his team of scientists have been studying the huntingtin protein for many years and have shared many important insights into how it works, and how it goes wrong in Huntington’s disease.
The N-terminal fragment
Huntingtin is a large protein, so studying all of it at once is difficult. After the gene was discovered in 1993, it was soon discovered that the most harmful bit of the mutant protein is near the very beginning. Scientists call this bit of huntingtin, containing the extra glutamines, ‘the N-terminal fragment’.
Besides the critical region that’s mutated in HD, this N-terminal fragment of the huntingtin protein has a number of other interesting features. Scientists have been studying it for nearly 20 years and are still finding a number of unexpected properties of this short piece of protein – properties that have big impact on cells.
Baggage handling in cells
Truant’s lab had already discovered that huntingtin’s first seventeen amino acid building blocks seem to be particularly important. They behave a bit like an ‘address label’ for the whole protein.
Depending on what’s happening to a cell, small chemical ‘tags’ can get attached to the first seventeen amino acids, or removed. The cell’s transport machinery can then read these tags like a bar code. Depending on the pattern of tags present, the huntingtin protein gets moved to different places in the cell. So, despite the fact that these seventeen amino acids are just a tiny portion of the whole huntingtin protein, they can have a huge impact on the protein’s location and function.
The adding and removing of these little tags is called ‘post-translational modification’. ‘Translation’ is the process of building proteins using the DNA instructions in our genes, so ‘post-translational’ just means ‘after the protein has been built’.
The magic numbers: thirteen and sixteen
“Specific tags on the thirteenth and sixteenth amino acids of huntingtin help determine whether or not it winds up in the nucleus.”
Understanding post-translational modification is really important in HD, because it seems to influence how harmful the protein is. For instance, in 2009, the group of William Yang at UCLA discovered that adding two of these tags prevented HD model mice from showing symptoms. Those tags were attached to the thirteenth and sixteenth amino acid building blocks of the huntingtin protein.
Since those two chemical tags seem crucial to how toxic the mutant huntingtin protein is, it’s important to understand them in more detail. Truant’s team set out to figure out the effect of those two tags on where the protein ended up, and how much harm it did once it was there.
What did they find out?
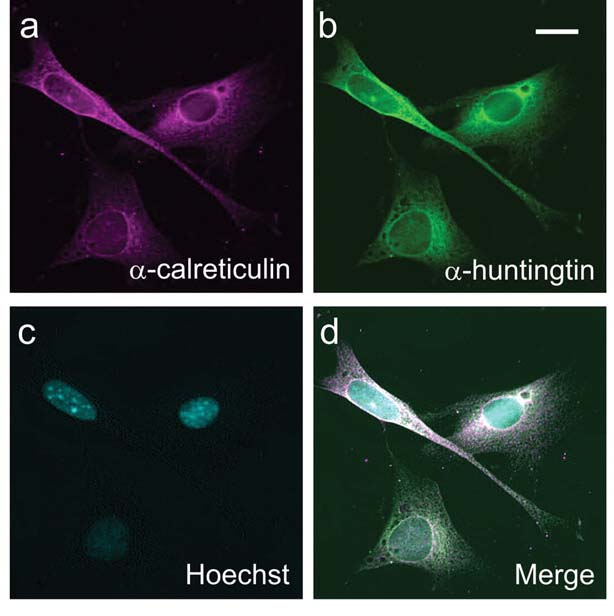
First, the researchers studied the protein in living cells, growing in a dish in the lab. They altered the cells genetically so that they just produced the first seventeen amino acid blocks of the huntingtin protein, which they called N17. The N17 huntingtin fragment was joined onto a jellyfish protein that glows yellow. That allows the N17 fragment to be watched as it moves around inside cells.
Truant’s team found that when the short fragment of huntingtin was tagged at the critical thirteenth and sixteenth spots, it was much more likely to end up in the nucleus of the cells.
The nucleus is a really important part of the cell – it’s where the DNA is stored, copied and read. Many researchers have suggested that huntingtin is more toxic to cells when it finds its way into the nucleus. But it can also do harm when it is outside the nucleus, so finding out where it’s most dangerous is important.
The team went further, showing that huntingtin is found in very specific locations within the nucleus. They also developed new techniques to look at just the tagged versions of the protein, rather than all the untagged copies floating around. These tools will be very useful for future studies investigating the role this tagging process plays in HD.
What about drugs?
This work shows that specific tags on the thirteenth and sixteenth amino acids of huntingtin help determine whether or not it winds up in the nucleus. It leads to the idea that drugs that alter the tagging of those amino acids might be able to reduce damage to cells.
How would a drug like that work? Or, in the language of drug hunters, what would its ‘target’ be?
Well, the chemical tags are attached to huntingtin by protein machines called ‘kinases’. And they’re removed by other machines called ‘phosphatases’. Kinases are a bit like staplers, while phosphatases are more like staple removers.
According to Truant’s work, drugs that reduce the activity of the kinase ‘stapler’ should reduce the tags on huntingtin, keeping it out of the nucleus. The next step would be to see where the protein ended up, and whether that made it more or less toxic.
To test this approach, Truant’s team used lots of different drugs that are known to target kinases. These ‘kinase inhibitor’ chemicals can help scientists to discover which particular kinase might be tagging their favorite protein. By blocking a particular kinase, then using the new tools they’ve developed to look for the tagged huntingtin, the researchers can discover which kinases are doing the tagging.
After testing drugs that block eighty different kinases, the team discovered that a particular kinase called ‘casein kinase 2’ – or ‘CK2’ – was particularly important. As expected, drugs that blocked CK2 caused huntingtin to be much less tagged at the crucial sites. They had the effect of keeping huntingtin out of the nucleus, instead targeting it to the ‘protein factory’ part of the cell, the endoplasmic reticulum or ‘ER’. In the process, the harm done to cells by the protein was increased.
Wait, they made the protein MORE toxic?!
Of course, a drug that makes huntingtin more toxic is the exact opposite of what we’d like to accomplish in people. So, don’t rush out to the drugstore and ask for a bottle of CK2 inhibitor.
If this work is going to lead to treatments for Huntington’s disease patients, we’ll want more tagged huntingtin, not less. At this early stage, though, Truant’s team was simply trying to figure out which kinase was the most powerful target.
By making things worse in cells with CK2-blocking drugs, they were able to confirm that CK2 is important in the tagging process we’re interested in. Future work can now be directed at understanding how achieve the reverse effect and make huntingtin less toxic.
This work, carried out in cells in a dish, is a long way from generating treatments that can be used in patients. But it’s an important step forward, that helps us understand one of the key players in the damage to cells in Huntington’s disease, and opens a new door for researchers to work on future treatments.
This article was updated on 27th June to clarify the effect of tagging and the CK2-blocking drugs.
Learn more
For more information about our disclosure policy see our FAQ…


